About our laboratory
The Multiscale Bioimaging and Bioinformatics Laboratory (MBB) is set up at both Biomedical Engineering Department of Tulane School of Science and Engineering and Biostatistics and Bioinformatics Department at Tulane School of Public Health and Tropical Medicine. In addition, we are part of Tulane Center for Bioinformatics and Genomics (CBG), which allow us to get access to rich resources at both schools.
The research dry labs of the CBG occupy ~750 sq ft, including two computer labs equipped with twelve computers, and a computer room housing two workstations. Software packages for biostatistics, bioinformatics, statistics and genetics, such as SAS and/or R, R Bioconductor, and Plink, are installed on these computers. The CBG has access to a controlled computer room housing our computing servers and cluster. The CBG has full access to the Biostatistics Data Center, which is housed in the Department of Biostatistics and Bioinformatics where Dr. Deng is the Chair, providing data entry and management services for researchers within and beyond the Tulane community. In addition, The CBG has access to the Tulane School of Public Health and Tropical Medicine (SPHTM) computer lab, which is located in the same building as that of CBG and is designed for training and education.
The research wet labs of the CBG occupy ~5,000 sq ft. The research labs house the major equipment needed for basic, clinical and translational research on musculoskeletal diseases, with the approaches of genomics, transciptomics, epigenomics and proteomics. The basic wet labs at CBG house the major equipment for experiments of, e.g., large scale genotyping, DNA re-sequencing, transcriptome gene expression profiling, proteomics, and molecular and cellular functional studies. DNA/RNA/protein extraction and cell isolations (e.g., isolating monocytes from peripheral blood samples) are routine procedures that are conducted in our research labs, as exemplified by our ongoing projects and our previous publications.
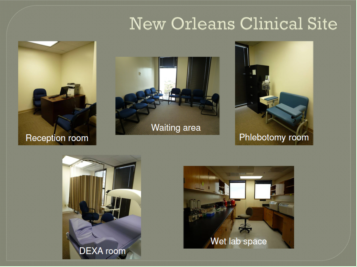
The CBG clinical recruitment site (shown in the photo) is located on the 11th floor of Tulane University School of Public Health Building, with a total area of 2,660 sq ft. This site has three full-time and five part-time employees (including study subject recruiters, phlebotomists, research assistants, and lab technicians). The clinical site houses an outpatient reception area, a waiting area, and multiple dedicated private interview offices for subject enrollment, exam rooms that are fully equipped for clinical examination, phenotyping and phlebotomy. The clinical recruitment site is within ~5 minute walking distance from the research wet labs of the CBG.
We have an exceptional good computing facility, either housed within our office building at the Tulane School of Public Health and Tropical Medicine or ready and convenient for us to access through intra- and internet at Tulane University and surrounding universities. We have > 50 desktops and >16 laptops (Intel Core 2 Duo and Quad Core CPUs 3-8 GB memory, 250-500 GB hard drives, MS Windows 7 Enterprise), two powerful computational servers (2-4 Intel/AMD Quad/twelve Core CPUs, 64-256 GB memory), and an in-house cluster (being able to run 210 jobs simultaneously). In particular, we recently acquired a Dell PowerEdge R910 Rack Server with four Intel Xeon E7540 six-core processors and 1 TB memory, which is ideal for handling and processing the next-generation sequencing-based genomic and epigenomic data. We have full access and have used the powerful Louisiana Optical Network Initiative, a state-of-the-art fiber optics network connecting Louisiana and Mississippi research universities and providing powerful distributed supercomputer resources. We are also frequent users of Tulane University High Performance Computing system, “Cypress", one of the fastest computers in the world. Cypress is the 3rd fastest among private American universities and 10th fastest among across all American universitities.
We have a large variety of software for daily operation, programming, data management, and biostatistics, statistical genetics, and bioinformatics analyses. Basic software packages for daily operation such as word processing, e-mail communication and web publishing, and for programming are in use on most desktop and laptop computers. Examples of these packages include Microsoft Office 2007/2010 Pro (e.g., Word for word processing, Excel for spreadsheet, and Powerpoint for presentation), Microsoft Access and MySQL (Database), and toolkit for print, web, and mobile publishing (Adobe Design Premium including Acrobat, Illustrator, InDesign, Photoshop, Dreamweaver, and Flash). Available programming languages in use include C, C++, Pascal, FORTRAN 77, Fortran 90, Perl/Bioperl, and Python/Biopython, which are often-employed for computational purposes, simulation studies in biostatistics and bioinformatics, and data handling.
General analytical packages such as SAS (Cary, NC), S-Plus (Insightful Corp., Seattle, WA), and R (http://www.r-project.org) are in use for routine biostatistical and bioinformatics analyses. They contain a variety of tools for routine analyses, such as evaluation of the appropriateness of analytical procedures (e.g., PROC UNIVARIATE in SAS for both graphical and analytic assessments of the normality of data), establishment of relationship between independent variables and dependent variables (e.g., linear regression by PROC REG in SAS, generalized linear regression including logistic regression by glm in R), and mixed model analyses (e.g., PROC MIXED in SAS). A variety of statistical and bioinformatical programs for analyzing genomic, functional genomic, epigenomic, and proteomic data are either regularly used or have been installed and ready for use.


